Chapter 5 Comparing Proportions
5.2 Facebook use example
In Chapter 9, we consider the following comparison of proportions example. A sample of students were asked their gender and the average number of times they visited Facebook in a day.
Of \(n_M\) males sampled, \(y_M\) had a high number of Facebook visits, and of \(n_F\) females sampled, \(y_F\) had a high number of visits.
Suppose the data is organized as a data frame as follows:
| Gender | Sample_size | Visits |
|---|---|---|
| male | \(n_M\) | \(y_M\) |
| female | \(n_F\) | \(y_F\) |
5.3 Sampling model
Suppose we have two independent samples where \(y_M\) is binomial(\(n_M, p_M\)) and \(y_F\) is binomial(\(n_F, p_F\)).
Write the proportions using a logistic model:
\[ \log\left(\frac{p}{1-p}\right) = \beta_0 + \beta_1 I(Gender = Male) \]
Note for females, the logit of \(p_F\) is given by \[ \log\left(\frac{p}{1-p}\right) = \beta_0 \] and for males the logit for \(p_M\) is given by \[ \log\left(\frac{p}{1-p}\right) = \beta_0 + \beta_1 \]
5.4 The data
Here’s the observed data:
5.5 Priors
In this model, \(\beta_0\) is the logit of the proportion of women who are high Facebook users and \(\beta_1\) represents the difference in the logits of the proportions for men and women.
Assume that you don’t know much about the location of \(\beta_0\), but you believe men and women are similar in their use of Facebook. So you assign a N(0, 31.6) prior to \(\beta_0\) with a high standard deviation, reflecting little knowledge. To reflect the belief that \(\beta_1\) is close to 0, you use a N(0, 0.71) prior.
The get_prior() function lists all parameters to define priors on for this particular model, assigning the result to prior. Then the two components of prior are assigned that reflect the statements above.
## prior class coef group resp dpar nlpar bound
## 1 b
## 2 b Gendermale
## 3 student_t(3, 0, 2.5) Intercept5.6 Posterior sampling
Here is the run of brm() where I use the prior specification in my_prior.
fit <- brm(family = binomial,
Visits | trials(Sample_Size) ~ Gender,
data = fb_data,
prior = my_prior,
iter = 1000,
refresh = 0)## Compiling Stan program...## Start samplingOne obtains the matrix of simulated values of the parameters by the posterior_samples() function.
## b_Intercept b_Gendermale lp__
## 1 0.149825204 -0.3929578 -10.89798
## 2 -0.077477237 -0.1550768 -10.36612
## 3 0.014914205 -0.2591160 -10.32287
## 4 -0.244104208 -0.2079800 -11.39275
## 5 -0.006092644 -0.2035093 -10.36111
## 6 0.012292753 -0.4538551 -10.53464The plot() function provides trace plots and density plots of each parameter.
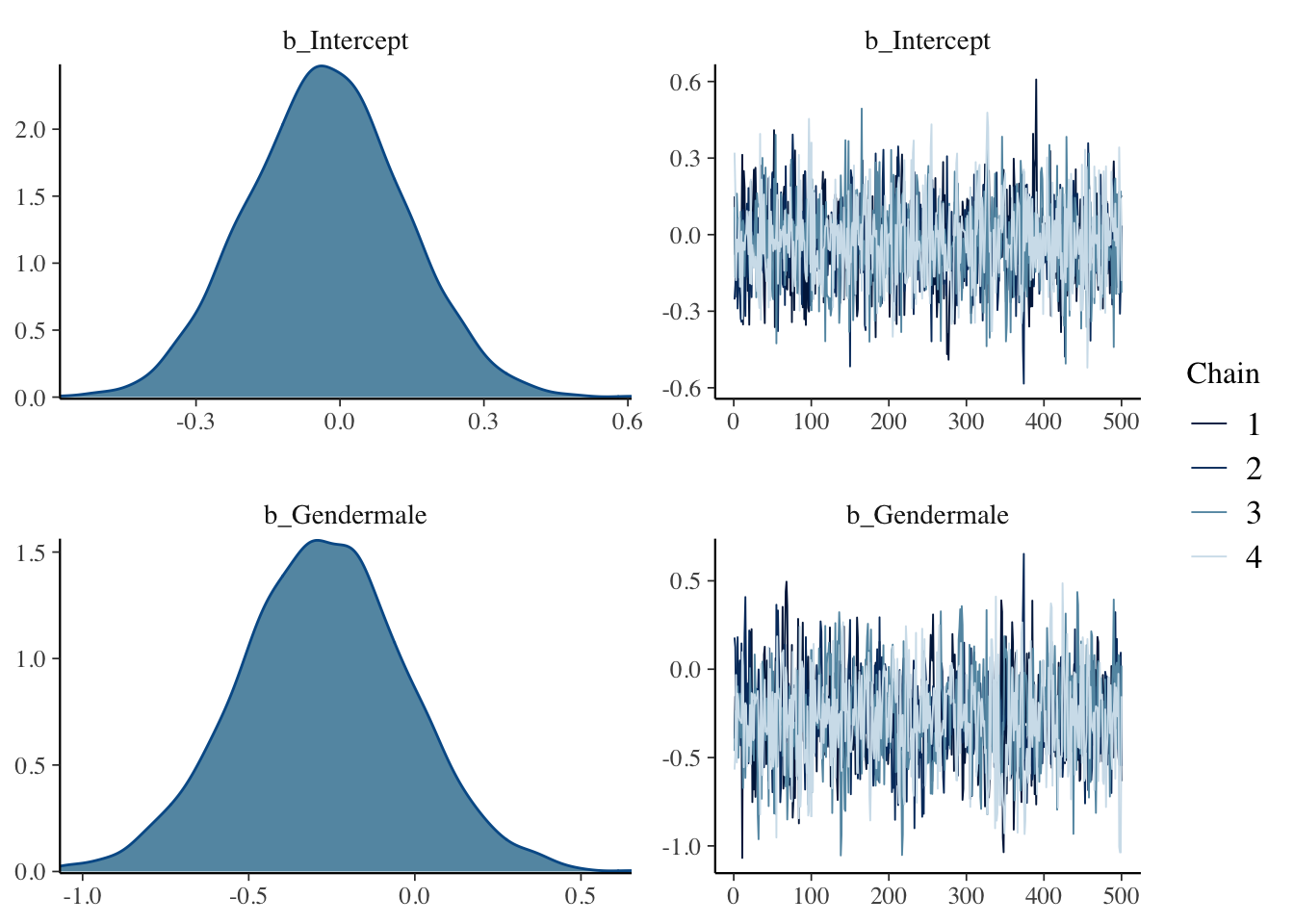
Posterior summaries are provided by the print() function.
## Family: binomial
## Links: mu = logit
## Formula: Visits | trials(Sample_Size) ~ Gender
## Data: fb_data (Number of observations: 2)
## Samples: 4 chains, each with iter = 1000; warmup = 500; thin = 1;
## total post-warmup samples = 2000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept -0.03 0.16 -0.34 0.28 1.00 2198 1576
## Gendermale -0.28 0.25 -0.78 0.23 1.01 1525 1121
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).