Chapter 6 Comparing Rates
6.2 Comparing two Poisson Rates
Suppose we observe two independent samples: \(x_1, ..., x_m\) are a random sample from a Poisson distribution with mean \(\lambda_x\), and \(w_1, ..., w_n\) are a random sample from a Poisson distribution with mean \(\lambda_y\). We are interested in learning about the ratio of Poisson means \[ \theta = \frac{\lambda_x}{\lambda_y} \]
6.3 Write as a log-linear model
Suppose we collect the observations
\[
y = c(x_1, ..., x_m, w_1, ..., w_n)
\]
and let group2 be an indicator variable for the second group.
\[
group2 = c(0, 0, ..., 0, 1, 1, ..., 1)
\]
Then we can represent the model as
\[
y_1, ..., y_{m+n}
\]
independent from Poisson distributions with means \(\lambda_1, ..., \lambda_{m_n}\) where the means follow the log-linear model
\[
\log \lambda_j = \beta_0 + \beta_1 group2
\]
In this model, \(\beta_0 = \log \lambda_x\), and \(\beta_0 + \beta_1 = \log \lambda_y\). So \(\beta_1 = \log(\lambda_y) - \log(\lambda_x)\) represents the increase in the means on the log scale.
6.4 The data
We collect web count visits for a number of days stored in the data frame web_visits in the ProbBayes package. The key variables are Day, the day of the week, and Count, the website visit count. We define a new variable Type that is either “weekend” or “weekday”.
We are interested in comparing the mean visit counts for weekdays and weekend days.
6.5 Priors
Here we are assume weakly informative priors on the regression parameters \(\beta_0\) and \(\beta_1\).
6.6 Bayesian fitting
## Compiling Stan program...## Start sampling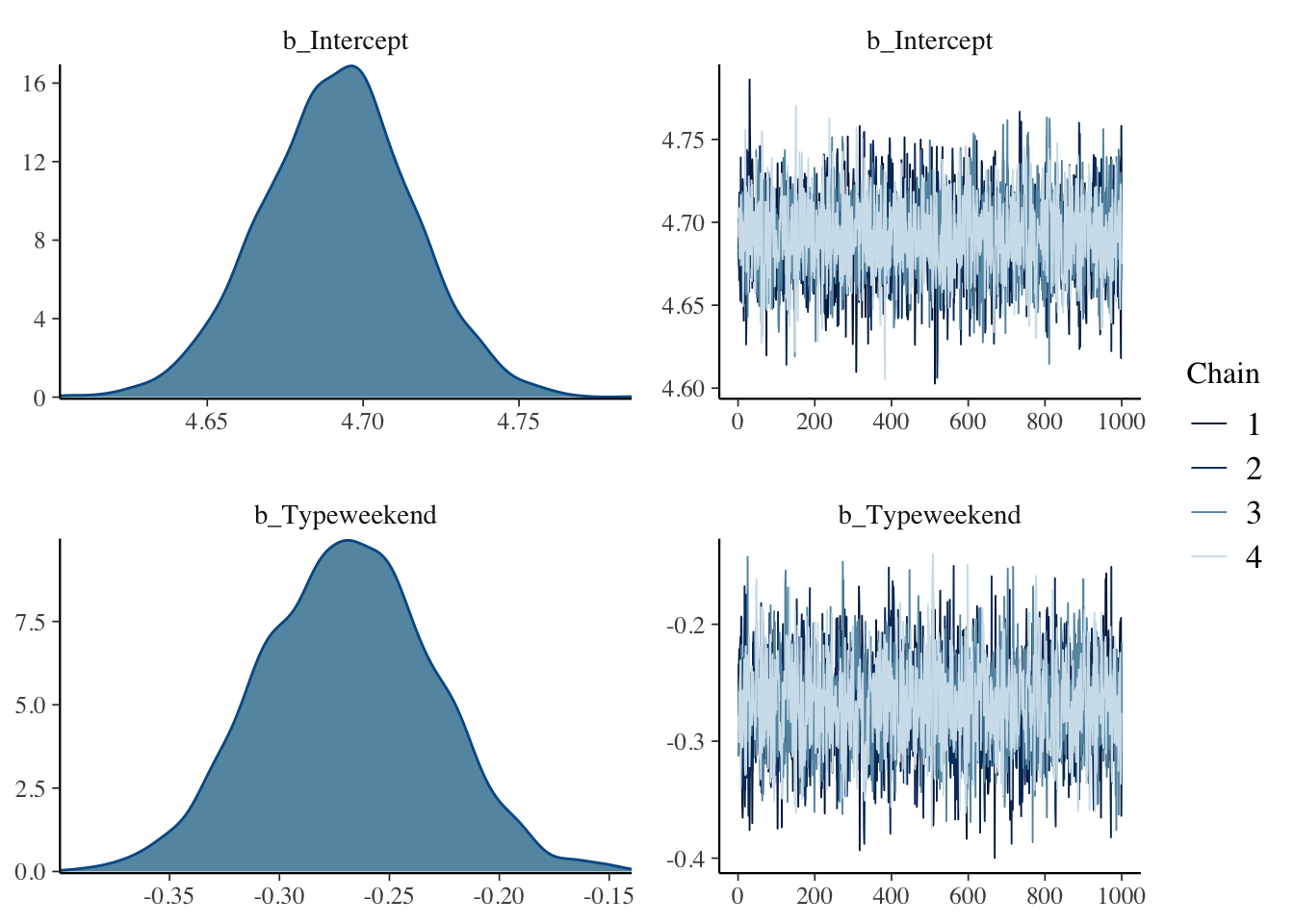
## Family: poisson
## Links: mu = log
## Formula: Count ~ Type
## Data: web_visits (Number of observations: 28)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 4.69 0.02 4.64 4.74 1.00 3888 2663
## Typeweekend -0.27 0.04 -0.35 -0.19 1.00 3292 2805
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).## b_Intercept b_Typeweekend lp__
## 1 4.683972 -0.3003928 -111.6071
## 2 4.690432 -0.2403990 -111.1721
## 3 4.665176 -0.2213930 -111.6114
## 4 4.683088 -0.2696015 -110.9016
## 5 4.678049 -0.2929583 -111.6562
## 6 4.699903 -0.2566505 -111.0721